Overview
The study of viral gene expression had led to the elucidation of several cellular and molecular processes operating in Eukaryotic cells. HIV is a very interesting study model as it exploits classical as well as non-conventional host pathways to accomplish efficient gene expression. As such, the viral transcripts that are generated by fully splicing of a precursor 9-kb viral mRNA associate with the classical cellular mRNP components and follow the same pathway as the host´s mRNAs. In contrast, other viral transcripts undergo partial splicing or are unspliced and therefore are expected to lack the main mRNP components being forced to exit the nucleus by an alternative pathway. In addition, viral transcripts exported through this pathway were shown to carry a trimethylated cap structure at their 5´ end, modification required for efficient gene expression but not detected to date in cellular mRNAs. In this career to produce high amount of viral proteins, HIV usurps several host proteins and/or machineries in its own benefit. The precise knowledge of all these processes is essential for the discovery and development of novel therapeutic targets for pharmaceutical intervention
Regulation of viral RNA metabolism by N6-methyladenosine (m6A)
In Progress
Gene expression from the HIV-1 genomic RNA
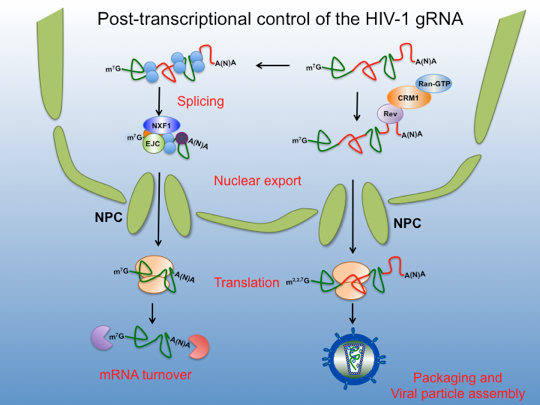
The study of viral gene expression had led to the elucidation of several cellular and molecular processes operating iFrom its birth during transcription to nuclear export, translation and decay, an mRNA molecule is always associated to a large number of proteins to form a messenger ribonucleoprotein complex (mRNP). It has been shown that the protein composition or nuclear imprinting acquired by a given mRNA can impact its cytoplasmic localization, translation and even its decay rates.
HIV transcription depends on the activity of the host RNAP II and, as a classical cellular mRNA, viral transcripts associate with several host RNA binding proteins in the nucleus. Interestingly, a single viral mRNA molecule, the so called full-length 9-kb unspliced genomic RNA (gRNA), is first processed by alternative splicing (fully or partially spliced transcripts) to give rise more than 40 transcripts coding for different regulatory and accessory proteins as well as the surface glycoproteins. However the unspliced form, the 9-kb gRNA, is also used as an mRNA coding for the major structural protein (Gag) and the proteins containing the viral enzymatic activities (Pol). This viral transcript exhibits features that are, a priori, not compatible with efficient gene expression including i) the presence of functional introns, an aspect incompatible with efficient nuclear export and translation; ii) AU-rich sequences that strongly interferes with gRNA stability, nuclear export and translation; iii) a 5’-untranslated region (5´-UTR) composed of several highly structured RNA motifs, which are expected to interfere with the ribosome recruitment by the canonical cap-dependent translation initiation mechanism and iv) a trimethylated cap structure that is expected to be inefficiently recognized by any of the canonical cellular cap-binding proteins associated to translation initiation. However, and despite all these constraints, the HIV-1 gRNA is efficiently exported to the cytoplasm by the viral protein Rev through the CRM1-RanGTP pathway to then reach the cytoplasm and recruit the host translational machinery giving rise to the large amounts of Gag that are required during viral replication. Once translated, the gRNA do not undergoes turnover as it is used as the genome that is incorporated into the viral particles. In the lab, we are mostly interested in study all these processes involved in the post-transcriptional control of the HIV-1 gRNA, specially on how nuclear events such as mRNP composition impact cytoplasmic localization and translation.
Viral and host factors involved in HIV-1 post-transcriptional control
The full-length 9-kb unspliced gRNA is used as the mRNA template to synthesize the major structural viral protein Gag and Pol, which contains the viral enzymes. This viral mRNA avoids splicing through a not well understood mechanism that is probably influenced by weak splice sites and assisted by the viral protein Rev.
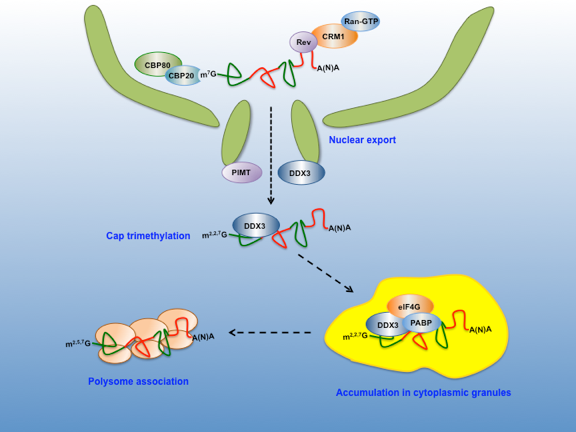
Despite the gRNA is expected to recruit co-transcriptionally the nuclear cap-binding complex (CBC, CBP20/CBP80) and other mRNP components, it may lack most of the classical cellular mRNP components such as the EJC and TAP/NXF1, which are recruited during the processing steps. As a consequence, the HIV-1 gRNA do not exits the nucleus by the canonical mRNA export pathway neither can benefit from the stimulating effects on nuclear export and translation provided by splicing through the recruitment of the EJC. Instead, the viral protein Rev binds to the RRE (Rev Responsive Element, an RNA motif present at the 3´ end of the gRNA and all partially spliced transcripts) and to CRM1 a karyopherin that shuttles between the nucleus and the cytoplasm and thus, exports the Rev/RRE complex as a cargo in a Ran-GTP-dependent manner.
Several cellular proteins including hRIP, eIF5A, Sam68 and the DEAD-box RNA helicases DDX1 and DDX3 have been shown to assist the Rev-mediated nuclear export yet, the mechanisms at play are poorly understood. One of the identified Rev partners is the methyltransferase, PIMT, which trimethylates the cap structure of the gRNA in a Rev-dependent manner. Interestingly, while the affinity of eIF4E and the CBC for a trimethylated cap is largely reduced resulting in the inhibition of translation, cap trimethylation was shown to increase Gag expression from the genomic RNA. Another Rev associated factor involved in gRNA translation is DDX3, which plays critical roles during the very early steps leading to ribosome recruitment and polysome association (see above).
As has been shown for cellular mRNAs, nuclear events are pivotal for the cytoplasmic destiny of the gRNA. Indeed, the viral protein Rev has been involved in cytoplasmic processes of the gRNA such as translation and packaging. Thus, we are investigating how nuclear host factors together with viral protein Rev are coupling nuclear export with translation initiation in order to gain insights into the molecular and cellular processes that regulate expression of this viral transcript, which is associated with non-canonical mRNP components.
Deciphering the atlas of the HIV-1 genomic mRNP
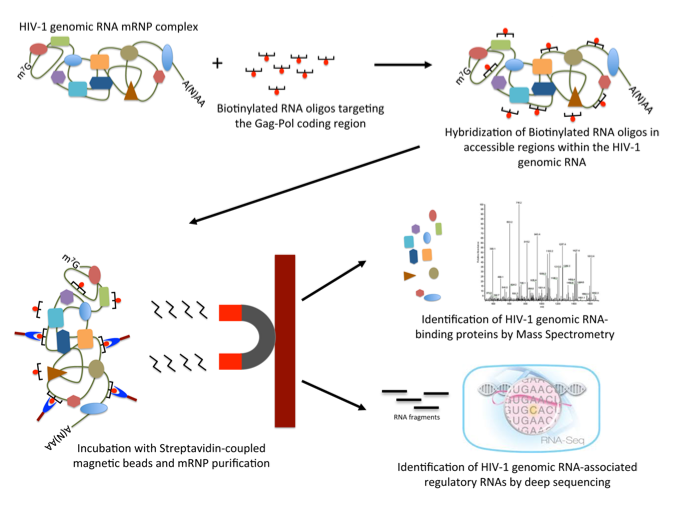
We are starting a new project aimed to isolate and characterize the whole components of the HIV-1 gRNA-containing mRNP. This project, funded by the Department of International Relationships (DRI) of Conicyt, will be carried out in tight collaboration with Drs. Melissa Moore and Emiliano Ricci from the University of Massachusetts Medical School.
The main goal of this project is to perform pull-downs of the HIV-1 gRNA (from nuclear and cytoplasmic factions, under different conditions, etc) and analyze both the proteins associated by Mass Spectrometry and the regulatory RNAs (miRNAs, lncRNAs, etc) by deep-sequencing.